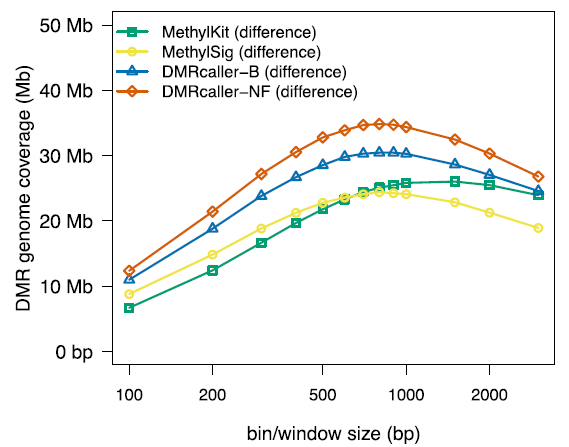
Description: DMRcaller is an R/Bioconductor package that computes Differentially Methylated Regions (DMRs) between two DNA methylation profiles obtained by Whole Genome Bisulfite Sequncing (WGBS). This tool, developed in collaboration with Dr. Radu Zabet, is highly configurable, user-friendly and optimized to detect statistically significant differences in DNA methylation also in poorly covered libraries. DMRcaller is able to perform most of genome wide analyses in a few hours, which represents a significant reduction in computing time over previous methods.
Reference: Catoni, M., Tsang, J.M.F., Greco, A.P., and Zabet, N.R. (2018). DMRcaller: a versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Research 46, e114.
Description: Pack-TYPE transposons are not autonomous DNA elements with the ability to shuffle exons across plant genomes. These elements are abundant in many plants (e.g. Pack-MULEs in rice) and we recently described their mobilization for the first time (Pack-CACTAs in Arabidopsis). We have developed the tool PackFinder, available through Bioconductor, to provide an automated and standardised workflow for the annotation and analysis of Pack-TYPE transposons. PackFinder can automatically identify previously manual annotated transposons with minimal false-postive rate and high confidence, and de novo annotate new elements in any genome reference. With minimal setup and expertise required, packFinder will facilitate the annotation and the study of novel Pack-TYPE elements.
Reference: Gisby J, Catoni M (2020). packFinder: de novo Annotation of Pack-TYPE Transposable Elements. R package version 1.1.0, https://github.com/jackgisby/packFinder.
Description: mobileRNA is an `R` package that provides a pipeline for the rapid identification of endogenous mobile RNA molecules in plant graft systems. The tool provides a pipeline for pre-processing and analysis of sRNA sequencing data, and soon mRNA sequencing data. It has been established that many different substances and molecules including RNAs can travel across the graft junction. Plant heterograft systems are comprised of two genotypes joined at the graft junction; hence, molecules produces and encoded by each genotype can move across the graft junction and be exchanged. These molecules could have implications to the regulation of gene expression and trait acquisition. Current methods utilise a step-wise mapping of samples to each genome within the graft system. While, here we introduce a new mapping method where we align each sample replicates to a merge genome reference comprised of both genome assemblies relating to the genotypes in the heterograft system.
Reference: Jeynes-Cupper K, Catoni M (2023). https://github.com/KJeynesCupper/mobileRNA